Bioinformatics
RNAseq, ChIPseq, ATACseq, CRISPR screening, single-cell, or something new!
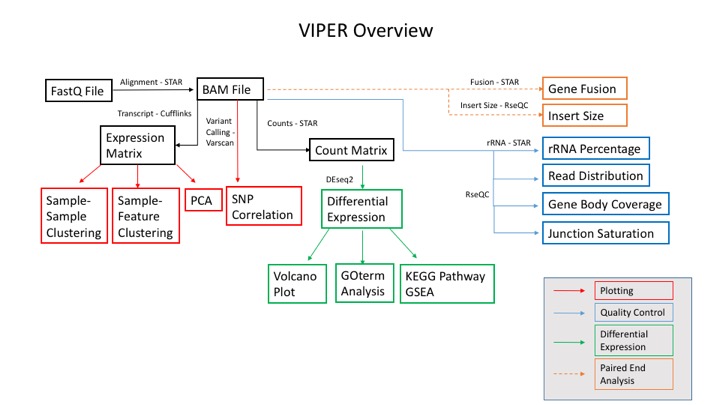
Visualization Pipeline for RNAseq (VIPER)
In collaboration with the Center for Functional Cancer Epigenetics (CFCE) at Dana-Farber Cancer Institute, the MBCF has developed a Visualization Pipeline for RNAseq (Viper) analysis tool.
Standard output includes:
-Aligned sorted.bam files
-Raw gene count and normalized FPKM matrices
-DESeq2 differential expression testing results
-Graphical Summary Report
ChIPseq analysis (ChiLin/CHIPS)
This pipeline was developed by the CFCE at Dana-Farber Cancer Institute. Reads are aligned with bowtie or BWA if paired-end followed by MACS for peak calling.
Output Includes:
-bigwig files for visualization
-peak list
-ChIPseq QC summary .pdf
Variant Calling
From WES, WGS, or custom panels
– BWA read alignment
– Variant calling by GATK
– Filtering by database and/or panel of normals
CRISPR screening
– Demultiplex staggered inline barcodes
– Align reads with bowtie to reference gRNA sequences
– Generation of count matrix
Copy Number Analysis from Ultra Low Pass Whole Genome Sequencing (ULP-WGS)
– Read alignment
– CNA by qDNAseq